Tuesday, 19.12.2017
The data set comes from 55000+ Song Lyrics, which contains over 55,000+ songs. It is a data frame with 55,000+ rows and four columns:
- artist
- song
- link
- text
library(igraph) library(ggraph) library(ggplot2) library(wordcloud2) library(dplyr) library(widyr) library(tidytext) library(tm) library(stringr) library(topicmodels) library(reshape2) library(quanteda) library(Rtsne) library(DT) library(knitr) library(animation) library(ldatuning) set.seed(201712) # Preperation ————————————————————- songs <– read.csv(“songdata.csv”) songs$song <– songs$song %>% as.character() songs$artist <– songs$artist %>% as.character() songs$link <– songs$link %>% as.character() songs$text <– songs$text %>% as.character()
Our goal is to perform a comprehensive analysis of the song texts to identify the Christmas songs. In order to do so, first we add an additional column to the data frame to give each song a label of either Christmas or Not Christmas, where every song which contains the words Christmas, Xmas or X-mas will be labeled as Christmas and otherwise as Not Christmas.
# Initialization of the Labels label <– character(dim(songs)[1]) for(i in 1:dim(songs)[1]){ if(str_detect(songs$song[i], “Christmas”) | str_detect(songs$song[i], “X-mas”) | str_detect(songs$song[i], “Xmas”)){ label[i] <– “Christmas” } else{ label[i] <– “Not Christmas” } } songs <– songs %>% mutate(Label = label)
This is just the initialization of the labels, later we will apply Naive Bayes to a training set to identify the other Christmas songs. First of all, we will start by exploring the data set by means of some intuitive descriptive approaches.
D3Vis <– function(edgeList, directed){ colnames(edgeList) <– c(“SourceName”, “TargetName”, “Weight”) # Min-Max & Inverse scaling, because the weights should represent distance/similarity edgeList$Weight <– 1 – edgeList$Weight weight.min <– edgeList$Weight %>% min weight.max <– edgeList$Weight %>% max edgeList$Weight <– (edgeList$Weight – weight.min)/(weight.max – weight.min) # Create a graph. Use simplyfy to ensure that there are no duplicated edges or self loops gD <– igraph::simplify(igraph::graph.data.frame(edgeList, directed=directed)) # Create a node list object (actually a data frame object) that will contain information about nodes nodeList <– data.frame(ID = c(0:(igraph::vcount(gD) – 1)), # because networkD3 library requires IDs to start at 0 nName = igraph::V(gD)$name) # Map node names from the edge list to node IDs getNodeID <– function(x){ which(x == igraph::V(gD)$name) – 1 # to ensure that IDs start at 0 } # And add them to the edge list edgeList <– plyr::ddply(edgeList, .variables = c(“SourceName”, “TargetName”, “Weight”), function (x) data.frame(SourceID = getNodeID(x$SourceName), TargetID = getNodeID(x$TargetName))) #+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++# # Calculate some node properties and node similarities that will be used to illustrate # different plotting abilities and add them to the edge and node lists # Calculate degree for all nodes nodeList <– cbind(nodeList, nodeDegree=igraph::degree(gD, v = igraph::V(gD), mode = “all”)) # Calculate betweenness for all nodes betAll <– igraph::betweenness(gD, v = igraph::V(gD), directed = directed) / (((igraph::vcount(gD) – 1) * (igraph::vcount(gD)–2)) / 2) betAll.norm <– (betAll – min(betAll))/(max(betAll) – min(betAll)) nodeList <– cbind(nodeList, nodeBetweenness=100*betAll.norm) # We are scaling the value by multiplying it by 100 for visualization purposes only (to create larger nodes) rm(betAll, betAll.norm) #Calculate Dice similarities between all pairs of nodes dsAll <– igraph::similarity.dice(gD, vids = igraph::V(gD), mode = “all”) F1 <– function(x) {data.frame(diceSim = dsAll[x$SourceID +1, x$TargetID + 1])} edgeList <– plyr::ddply(edgeList, .variables=c(“SourceName”, “TargetName”, “Weight”, “SourceID”, “TargetID”), function(x) data.frame(F1(x))) rm(dsAll, F1, getNodeID, gD) #++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++# # We will also create a set of colors for each edge, based on their dice similarity values # We’ll interpolate edge colors based on the using the “colorRampPalette” function, that # returns a function corresponding to a collor palete of “bias” number of elements (in our case, that # will be a total number of edges, i.e., number of rows in the edgeList data frame) F2 <– colorRampPalette(c(“#FFFF00”, “#FF0000”), bias = nrow(edgeList), space = “rgb”, interpolate = “linear”) colCodes <– F2(length(unique(edgeList$diceSim))) edges_col <– sapply(edgeList$diceSim, function(x) colCodes[which(sort(unique(edgeList$diceSim)) == x)]) rm(colCodes, F2) #++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++# # revise transformation of the weights edgeList$Weight <– –(edgeList$Weight*(weight.max – weight.min) + weight.min – 1) # Let’s create a network D3_network_LM <– networkD3::forceNetwork(Links = edgeList, # data frame that contains info about edges Nodes = nodeList, # data frame that contains info about nodes Source = “SourceID”, # ID of source node Target = “TargetID”, # ID of target node Value = “Weight”, # value from the edge list (data frame) that will be used to value/weight relationship amongst nodes NodeID = “nName”, # value from the node list (data frame) that contains node description we want to use (e.g., node name) Nodesize = “nodeBetweenness”, # value from the node list (data frame) that contains value we want to use for a node size Group = “nodeDegree”, # value from the node list (data frame) that contains value we want to use for node color # height = 500, # Size of the plot (vertical) # width = 1000, # Size of the plot (horizontal) fontSize = 20, # Font size linkDistance = networkD3::JS(“function(d) { return 10*d.value; }”), # Function to determine distance between any two nodes, uses variables already defined in forceNetwork function (not variables from a data frame) # linkWidth = networkD3::JS(“function(d) { return d.value/5; }”),# Function to determine link/edge thickness, uses variables already defined in forceNetwork function (not variables from a data frame) opacity = 0.85, # opacity arrows = directed, zoom = TRUE, # ability to zoom when click on the node # opacityNoHover = 0.1, # opacity of labels when static legend = F, linkColour = edges_col) # edge colors # Plot network D3_network_LM } </code><a id=“user-content-exploration-the-initial-christmas-songs” class=“anchor” href=“https://github.com/eodaGmbH/christmas_songs/blob/master/notebook.Rmd#exploration-the-initial-christmas-songs” aria–hidden=“true”></a><a id=“user-content-cleaning–tokenization” class=“anchor” href=“https://github.com/eodaGmbH/christmas_songs/blob/master/notebook.Rmd#cleaning–tokenization” aria–hidden=“true”></a>
We should start with the data cleaning and tokenization. Afterwards, the Christmas songs will be selected and saved as a variable.
songs.unnest <– songs %>% unnest_tokens(word, text) %>% anti_join(tibble(word = stop_words$word)) %>% filter(!str_detect(word, “\\d+”)) xmas.unnest <– songs.unnest %>% filter(Label == “Christmas”) </code><a id=“user-content-correlation-analysis” class=“anchor” href=“https://github.com/eodaGmbH/christmas_songs/blob/master/notebook.Rmd#correlation-analysis” aria–hidden=“true”></a>
Now we can start analyzing the initial Christmas songs by means of correlations from different perspectives. In the following, we will visualize the correlations with the networkD3 html widget where nodes with the same total number of connections will be given the same color and the color of the edge implies the number of common neighbors shared by two nodes. Moreover, the size of a node indicates the centrality of it, which is defined by the betweenness, i.e. the number of shortest paths going through it. Where the distance between two nodes is the minimum maximum transformation of 1 minus the correlation, which makes sense because intuitively the higher the correlation, the nearer two nodes should be. Moreover, the shorter the distance, the wider the edge.
Note that the correlations are always based on lyrics.
The correlation between words which appeared more than 100 times and are correlated with at least one other word with a correlation greater than 0.55.
correlation.words <– xmas.unnest %>% group_by(word) %>% filter(n() > 100) %>% ungroup() %>% pairwise_cor(word, song, sort = T) # Network visualization correlation.words %>% filter(correlation > 0.55) %>% D3Vis(directed = F) </code><a id=“user-content-correlation-between-songs” class=“anchor” href=“https://github.com/eodaGmbH/christmas_songs/blob/master/notebook.Rmd#correlation-between-songs” aria–hidden=“true”></a>
The correlation between songs which are correlated with at least 3 other songs with a correlation greater than 0.75. With this, we may detect similiar or just slightly modified songs.
correlation.songs <– xmas.unnest %>% pairwise_cor(song, word, sort = T) # Network visualization correlation.songs %>% filter(correlation > 0.75) %>% group_by(item1) %>% filter(n() >= 3) %>% ungroup() %>% D3Vis(directed = F) </code><a id=“user-content-correlation-between-certain-words” class=“anchor” href=“https://github.com/eodaGmbH/christmas_songs/blob/master/notebook.Rmd#correlation-between-certain-words” aria–hidden=“true”></a>
The correlation between certain words
correlation.words %>% filter(item1 == “christus” | item1 == “jesus” | item1 == “snow” | item1 == “reindeer” | item1 == “home” | item1 == “holy” | item1 == “love” | item1 == “tree” | item1 == “white” | item1 == “christmas”, correlation > 0.4) %>% D3Vis(directed = F) </code><a id=“user-content-correlation-between-artists” class=“anchor” href=“https://github.com/eodaGmbH/christmas_songs/blob/master/notebook.Rmd#correlation-between-artists” aria–hidden=“true”></a>
The correlation between artists
correlation.artists <– xmas.unnest %>% pairwise_cor(artist, word, sort = T) # Network Visualization correlation.artists %>% filter(correlation > 0.8) %>% group_by(item1) %>% filter(n() >= 3) %>% ungroup() %>% D3Vis(directed = F) </code><a id=“user-content-word-cloud” class=“anchor” href=“https://github.com/eodaGmbH/christmas_songs/blob/master/notebook.Rmd#word-cloud” aria–hidden=“true”></a>
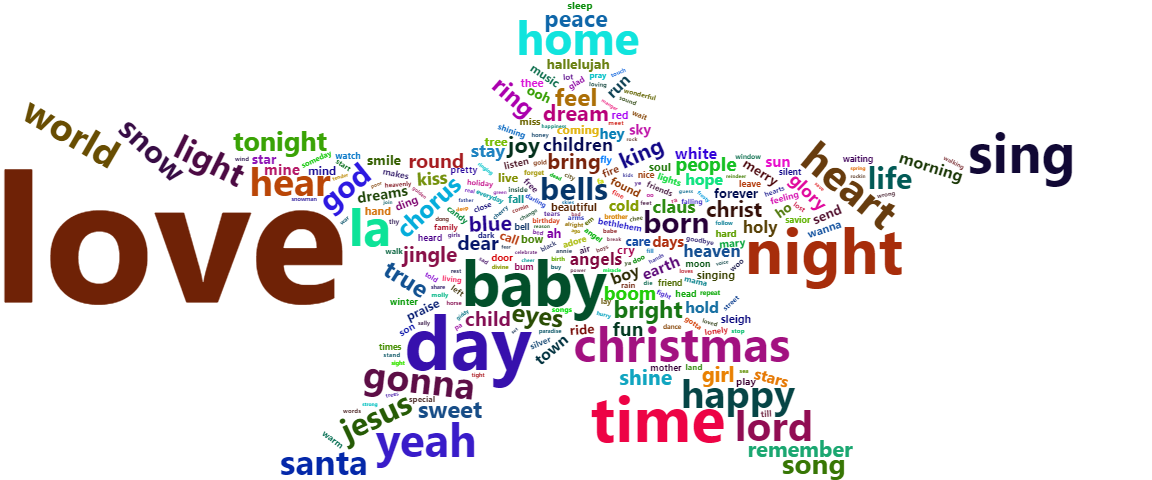
Word cloud of the initial Christmas songs
xmas.cloud <– xmas.unnest %>% count(word) %>% as.data.frame() xmas.cloud %>% wordcloud2(minSize = 3, shape = ‘star’) </code><a id=“user-content-naive-bayes” class=“anchor” href=“https://github.com/eodaGmbH/christmas_songs/blob/master/notebook.Rmd#naive-bayes” aria–hidden=“true”></a>
Naive Bayes is a popular supervised machine learning algorithm to handle classification problems with a huge amount of features. It is „naive“ in the sense that, conditioned on a class, the features are assumed to be independently distributed. In our case, we would like to know, given a bunch of features, i.e. the tf-idf of words in a document, whether a song should be classified as Christmas song or not by Naive Bayes.
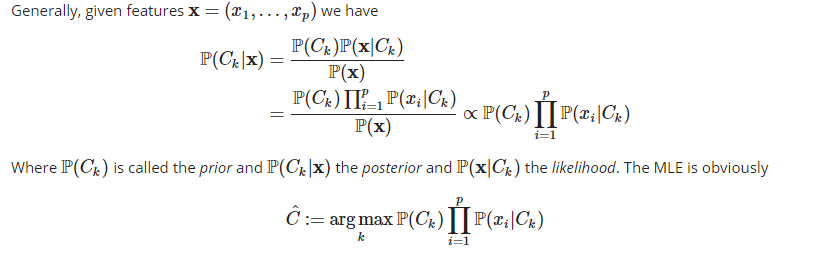
The harder part of constructing the maximum likelihood estimator is the choice of the prior distribution, i.e. the probability distribution of the classes. Where it is usually assumed to be uniformly distributed or estimated by the class frequencies. In our case the multinomial distribution for the likelihood and the uniform distribution for the prior are used, which means we have no prejudice regarding the categorization of the songs without given further information.
# Document Feature Matrix songs.dfm.tfidf <– corpus(songs, text_field = “text”, docid_field = “song”) %>% dfm(tolower = T, stem = TRUE, remove_punct = TRUE, remove = stopwords(“english”)) %>% dfm_trim(min_count = 5, min_docfreq = 3) %>% dfm_weight(type = “tfidf”) # Determine the Indizes for the training set christmas.index <– which(label == “Christmas”) not_christmas.index <– which(label == “Not Christmas”) christmas.train.index <– christmas.index not_christmas.train.index <– sample(not_christmas.index, length(christmas.index)) train.index <– c(christmas.train.index, not_christmas.train.index) label.train <– label[train.index] trainning.set <– songs.dfm.tfidf[train.index, ] # Train the Model classifier_NB <– textmodel_NB(trainning.set, label.train) # Prediction predictions <– classifier_NB %>% predict(newdata = songs.dfm.tfidf) # Confusion Matrix confusion <– table(predictions$nb.predicted, label) confusion
So we have identified 2965 hidden Christmas songs and there are 2 songs out of the initial 500 Christmas songs that are rejected by Naive Bayes as Christmas songs.
Explore the hidden Christmas songs
# Document Feature Matrix songs.dfm.tfidf <– corpus(songs, text_field = “text”, docid_field = “song”) %>% dfm(tolower = T, stem = TRUE, remove_punct = TRUE, remove = stopwords(“english”)) %>% dfm_trim(min_count = 5, min_docfreq = 3) %>% dfm_weight(type = “tfidf”) # Determine the Indizes for the training set christmas.index <– which(label == “Christmas”) not_christmas.index <– which(label == “Not Christmas”) christmas.train.index <– christmas.index not_christmas.train.index <– sample(not_christmas.index, length(christmas.index)) train.index <– c(christmas.train.index, not_christmas.train.index) label.train <– label[train.index] trainning.set <– songs.dfm.tfidf[train.index, ] # Train the Model classifier_NB <– textmodel_NB(trainning.set, label.train) # Prediction predictions <– classifier_NB %>% predict(newdata = songs.dfm.tfidf) # Confusion Matrix confusion <– table(predictions$nb.predicted, label) confusion
# Correlation hidden.correlation.words <– hidden.unnest %>% group_by(word) %>% filter(n() > 15) %>% ungroup() %>% pairwise_cor(word, song, sort = T) # Network visualization hidden.correlation.words %>% filter(correlation > 0.65) %>% group_by(item1) %>% filter(n() >= 20) %>% ungroup() %>% D3Vis(directed = F)
We have therefore successfully identified a bunch of religous christmas songs, whose titles usually do not contain the word „Christmas“ or „X-mas“.
Latent Dirichtlet Allocation & t-Statistics Stochastic Neighbor Embedding
Data Preparation
Only the top 300 features for the Christmas songs including the hidden ones will be used to calculate the Rtsne & LDA, else the memory space will not be sufficient.
xmas.dfm.tfidf <– songs.dfm.tfidf %>% dfm_subset(Label == “Christmas” | Label == “Hidden Christmas”) songs.dfm.tfidf_300 <– songs.dfm.tfidf %>% dfm_select(pattern = xmas.dfm.tfidf %>% topfeatures(300) %>% names(), selection = “keep”) xmas.dfm.tfidf_300 <– xmas.dfm.tfidf %>% dfm_select(pattern = xmas.dfm.tfidf %>% topfeatures(300) %>% names(), selection = “keep”)
LDA
LDA stands for Latent Dirichtlet Allocation, which was introduced in Blei, Ng, Jordan (2003). It is a generative probabilistic model of a corpus, where the documents are represented as random mixtures over latent topics and for a single document there are usually only a few topics that are assigned unneglectable probabilities. Moreover, each topic is characterized by a distribution over words, where usually only a small set of words will be assigned significant probabilities for a certain topic. Either the variational expectation maximization algorithm or Gibbs sampling is used for the statistical inference of the parameters.
LDA requires a fixed number of topics, i.e. it assumes that the number of topics should already be known before applying the algorithm. However, there are possibilities to determine the optimal number of topics by different performance metrics, see Nikita, by using the package ldatuning.
OptimalNumber <– FindTopicsNumber(LDA_xmas <– xmas.dfm.tfidf_300 %>% convert(to = “topicmodels”), topics = seq(2, 8, by = 1), mc.cores = 2, metrics = c(“CaoJuan2009”, “Arun2010”, “Deveaud2014”), method = “VEM”, verbose = T)
FindTopicsNumber_plot(OptimalNumber
Therefore, we will choose 8 as the optimal number of topics.
LDA_xmas <– xmas.dfm.tfidf_300 %>% convert(to = “topicmodels”) %>% LDA(k = 8)
We may use the package tidytext to inspect the topic probability distribution of each document, i.e. for each document the sum of the probabilities that it belongs to a topic from 1 to 8 is equal to 1.
LDA_xmas %>% tidy(matrix = “gamma”) %>% datatable(rownames = F)
Analogously, we can also obtain the probability distribution of words for each topic, i.e. for each topic the sum of probabilities that it generates different words is equal to 1.
LDA_xmas %>% tidy(matrix = “beta”) %>% datatable(rownames = F)
The top terms for each topic are:
# LDA for the Christmas songs terms(LDA_xmas, 10)
t-SNE
Developed by van der Maaten and Hinton (2008), t-SNE stands for t-Statistics Stochastic Neighborhood Embedding, which is a dimensionality reduction technique that is formulated to capture the local clustering structure of the original data points. It is non-linear and non-deterministic.
The following computation will take about 30 minutes.
# t-Statistics Stochastic Neighbor Embedding ——————————– index.unique.songs <– !songs.dfm.tfidf_300 %>% as.matrix() %>% duplicated() songs.unique <– songs.dfm.tfidf_300[index.unique.songs, ] %>% as.matrix() tsne.all <– Rtsne(songs.unique) songs_2d <– tsne.all$Y %>% as.data.frame() %>% mutate(Label = label[index.unique.songs])
songs_2d %>% ggplot(aes(x = V1, y = V2, color = Label)) + geom_point(size = 0.25) + scale_color_manual(values = c(“Not Christmas” = “#a6a6a6”, “Christmas” = “#88ab33”, “Hidden Christmas” = “#F98948”, “Hidden Not Christmas” = “#437F97”)) + guides(color = guide_legend(override.aes = list(size = 5))) + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), panel.background = element_rect(fill = “black”))
What if we repeat the procedure for more than one iteration?
So far we have only run the Naive Bayes for one iteration. However, we may repeat this procedure for more than one iteration, i.e. train a Naive Bayes classifier and relabel all the false positives as Hidden Christmas/Christmas and all the false negatives as Hidden Not Christmas/Not Christmas over and over again.
First of all, we prepare the data again to avoid bugs.
songs <– read.csv(“songdata.csv”) songs$song <– songs$song %>% as.character() songs$artist <– songs$artist %>% as.character() songs$link <– songs$link %>% as.character() songs$text <– songs$text %>% as.character() # Initialization of the Labels label <– character(dim(songs)[1]) for(i in 1:dim(songs)[1]){ if(str_detect(songs$song[i], “Christmas”) | str_detect(songs$song[i], “X-mas”) | str_detect(songs$song[i], “Xmas”)){ label[i] <– “Christmas” } else{ label[i] <– “Not Christmas” } } songs <– songs %>% mutate(Label = label) songs.dfm.tfidf <– corpus(songs, text_field = “text”, docid_field = “song”) %>% dfm(tolower = T, stem = TRUE, remove_punct = TRUE, remove = stopwords(“english”)) %>% dfm_trim(min_count = 5, min_docfreq = 3) %>% dfm_weight(type = “tfidf”) results <– data.frame(precision = numeric(10), recall = numeric(10), f1_score = numeric(10))
Run 10 iterations.
for(i in 1:10){ # Determine the Indizes christmas.index <– which(label == “Christmas”) not_christmas.index <– which(label == “Not Christmas”) if(length(christmas.index) < length(not_christmas.index)){ christmas.train.index <– christmas.index not_christmas.train.index <– sample(not_christmas.index, length(christmas.index)) } else{ not_christmas.train.index <– not_christmas.index christmas.train.index <– sample(christmas.index, length(not_christmas.index)) } train.index <– c(christmas.train.index, not_christmas.train.index) label.train <– label[train.index] trainning.set <– songs.dfm.tfidf[train.index, ] # Train the Model classifier_NB <– textmodel_NB(trainning.set, label.train) # Prediction predictions <– classifier_NB %>% predict(newdata = songs.dfm.tfidf) # Confusion Matrix confusion <– table(predictions$nb.predicted, label) precision <– confusion[1, 1]/sum(confusion[1, ]) recall <– confusion[1, 1]/sum(confusion[, 1]) f1_score <– 2*precision*recall/(precision + recall) # The hidden (not) Christmas Songs ———————————————- hidden.index <– (predictions$nb.predicted == “Christmas”) & (songs$Label == “Not Christmas”) hidden_not.index <– (predictions$nb.predicted == “Not Christmas”) & (songs$Label == “Christmas”) hidden.xmas <– songs[hidden.index, ] hidden.not_xmas <– songs[hidden_not.index, ] label[hidden.index] <– “Hidden Christmas” label[hidden_not.index] <– “Hidden Not Christmas” songs_2d <– tsne.all$Y %>% as.data.frame() %>% mutate(Label = label[index.unique.songs]) random.forest <– songs_2d %>% ggplot(aes(x = V1, y = V2, color = Label)) + geom_point(size = 0.25) + scale_color_manual(values = c(“Not Christmas” = “#a6a6a6”, “Christmas” = “#88ab33”, “Hidden Christmas” = “#F98948”, “Hidden Not Christmas” = “#437F97”)) + guides(color = guide_legend(override.aes = list(size = 5))) + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), panel.background = element_rect(fill = “black”)) + ggtitle(paste(“Iteration:”, i)) # Change the labels label[hidden.index] <– “Christmas” label[hidden_not.index] <– “Not Christmas” songs$Label <– label songs.dfm.tfidf@docvars$Label <– label results[i, ] <– c(precision, recall, f1_score) plot(random.forest) }
results %>% mutate(index = 1:10) %>% melt(id = “index”) %>% ggplot(aes(x = index, y = value, color = variable)) + geom_line()
Then the precision as well as the f1 score grow monotonically at first and then converge to a value around 0.95, which means there are not many „Hidden Christmas Songs“ and „Hidden Not Christmas Songs“ left to be detected. However, in this procedure we always believe that the Naive Bayes classifier is 100% accurate, which is hardly possible. Thus, in each iteration there are some songs falsely classified by Naive Bayes as „Christmas“, which will be used in the next iteration in the training set to train the Naive Bayes classifier. With this accumulating error we might have the apprehension that the results are actually worse with more iterations.
confusion
At the end we have roughly half of the songs classified as „Christmas“ and the other half as „Not Christmas“, which seems very implausible. It raises the question whether or not there is an optimal number of iterations, however, we simply can not manually control whether all the 57,650 songs are correctly classified or not. This remains an open question to be answered.
Get started now:
We look forward to engaging with you.