
10 Tidyverse functions that might save your day
In this blog post, we will present 10 Tidyverse functions that are often overlooked by beginners but have proven to be very useful in the right context. We will first describe a problem that we faced in practice in a similar form and then explain how Tidyverse helps us to solve this problem.

10 Tidyverse functions that might save your day
In this blog post, we will present 10 Tidyverse functions that are often overlooked by beginners but have proven to be very useful in the right context. We will first describe a problem that we faced in practice in a similar form and then explain how Tidyverse helps us to solve this problem.
For the preparation and analysis of data in R, the Tidyverse packages have become an industry standard in the last few years. At eoda, we use many features from the Tidyverse to increase the efficiency of our daily work.
Content
- crossing
- rowwise
- pluck
- rownames_to_column & rowid_to_column
- parse_number
- fct_lump_*
- fct_reorder + geom_col
- separate & separate_rows
- str_flatten_comma
- arrange + distinct
- Conclusion
As expected, we start loading the necessary libraries:
library(tidyverse)1. crossing
Problem:
For the first example, we consider a statistical application. Given two vectors of numerical means and standard deviations, we want to collect all the combination of values that occur in a data frame.
Solution:
The crossing() function from the tidyr package serves exactly this purpose. It takes an arbitrary number of vectors as input and builds all possible combinations of the occurring values:
means <- c(-1, 0, 1)
standard_deviations <- c(0.1, 0.5, 1)
mean_sd_combinations <- crossing(means, standard_deviations)
mean_sd_combinationsCode language: R (r)| means | standard_deviations |
|---|---|
| -1 | 0.1 |
| -1 | 0.5 |
| -1 | 1.0 |
| 0 | 0.1 |
| 0 | 0.5 |
| 0 | 1.0 |
| 1 | 0.1 |
| 1 | 0.5 |
| 1 | 1.0 |
Bonus:
crossing() can take not only vectors but also data frames as input. In this case all combinations of the rows are formed.
This is especially useful if one of the data frames provides “global” information (in the following example population_data), which is valid for all observations, and the second data frame provides “local” information, which differs between observations or groups (in the example group_data).
population_data <- tibble(
global_feature_1 = "e",
global_feature_2 = 5,
)
population_dataCode language: R (r)| global_feature_1 | global_feature_2 |
|---|---|
| e | 5 |
group_data <- tibble(
group = 1:3,
local_feature_1 = c(2, 5, 3),
local_feature_2 = c(TRUE, FALSE, FALSE)
)
group_dataCode language: R (r)| group | local_feature_1 | local_feature_2 |
|---|---|---|
| 1 | 2 | TRUE |
| 2 | 5 | FALSE |
| 3 | 3 | FALSE |
As a result of crossing(), we get a single data frame in which each row contains both the global and the group-specific values:
crossing(population_data, group_data)| global_feature_1 | global_feature_2 | group | local_feature_1 | local_feature_2 |
|---|---|---|---|---|
| e | 5 | 1 | 2 | TRUE |
| e | 5 | 2 | 5 | FALSE |
| e | 5 | 3 | 3 | FALSE |
2. rowwise
Problem:
We stay with the application example from Section 1. For each of the mean-standard deviation combinations, five random values (samples) of a standard normal distribution are to be drawn and added to the data frame in a new column.
Consequently, we need to act at the row level here: Each row of the Data Frame forms a related unit. The newly generated values of the first row are based solely on the remaining values of the first row.
Another peculiarity is that we add multiple entries per cell, not just a single one. In order for this to be compatible with the structure of a data frame, they must be combined into a list. Consequently, the new column is a list column – a column consisting of lists.
Solution:
One way is to use the map() family from the purrr package. The means and standard_deviations columns, to which the rnorm() function is applied, are referenced by the .x and .y placeholders:
random_samples_map <- mean_sd_combinations |> mutate(
samples = map2(means, standard_deviations, ~ rnorm(n = 5, mean = .x, sd = .y))
)
random_samples_map |> head()
<em>## # A tibble: 6 × 3</em>
<em>## means standard_deviations samples </em>
<em>## <dbl> <dbl> <list> </em>
<em>## 1 -1 0.1 <dbl [5]></em>
<em>## 2 -1 0.5 <dbl [5]></em>
<em>## 3 -1 1 <dbl [5]></em>
<em>## 4 0 0.1 <dbl [5]></em>
<em>## 5 0 0.5 <dbl [5]></em>
<em>## 6 0 1 <dbl [5]></em>Code language: R (r)Each entry of the new samples column consists of a list of five drawn values from a standard normal distribution:
random_samples_map$samples[[1]]
<em>## [1] -1.0416796 -0.9907691 -0.9249944 -0.8859866 -1.0676741</em>Code language: R (r)For many use cases the rowwise() function from the dplyr package offers a more user-friendly alternative. The column names means and standard_deviations can be used here directly in the call to the rnorm() function without the use of wildcards.
Since the new column consists of lists, the call to rnorm() must be made within list():
random_samples_map <- mean_sd_combinations |>
dplyr::rowwise() |>
mutate(samples = list(rnorm(n = 5, mean = means, sd = standard_deviations)))
random_samples_map$samples[[1]]
<em>## [1] -0.9437694 -0.9311953 -1.0259749 -1.0115392 -1.0614477</em>Code language: R (r)Bonus:
When working with ‘list columns‘ the dplyr function nest_by() can be very useful, which unlike tidyr::nest() forms groups line by line.
As an example, we form a separate group for each cyl (cylinder) value from the mtcars dataset. All remaining mtcars columns are bundled into a new column consisting of data frames.
mtcars |> nest_by(cyl)
<em>## # A tibble: 3 × 2</em>
<em>## # Rowwise: cyl</em>
<em>## cyl data</em>
<em>## <dbl> <list<tibble[,10]>></em>
<em>## 1 4 [11 × 10]</em>
<em>## 2 6 [7 × 10]</em>
<em>## 3 8 [14 × 10]</em>Code language: R (r)From this, we can add a new column with linear models of mpg (miles per gallon) as a function of hp (horse power).
In a last step, we extract from this the slope coefficients, one number per cylinder value. The result is a single data frame containing the original data, the model objects and the slope coefficients:
mtcars |>
nest_by(cyl) |>
mutate(model = list(lm(mpg ~ hp, data = data))) |>
mutate(slope = coef(model)[2])
<em>## # A tibble: 3 × 4</em>
<em>## # Rowwise: cyl</em>
<em>## cyl data model slope</em>
<em>## <dbl> <list<tibble[,10]>> <list> <dbl></em>
<em>## 1 4 [11 × 10] <lm> -0.113 </em>
<em>## 2 6 [7 × 10] <lm> -0.00761</em>
<em>## 3 8 [14 × 10] <lm> -0.0142</em>Code language: R (r)3. pluck
Problem:
From the nested list l, we want to select the string “c” of the lowest level, i.e., the third value of element b in the first list element of a. In total, we have to extract a value from the fourth level of the list.
l <- list(a = list(c(1, 2, list(b = c("a", "b", "c")))))
l
<em>## $a</em>
<em>## $a[[1]]</em>
<em>## $a[[1]][[1]]</em>
<em>## [1] 1</em>
<em>## </em>
<em>## $a[[1]][[2]]</em>
<em>## [1] 2</em>
<em>## </em>
<em>## $a[[1]]$b</em>
<em>## [1] "a" "b" "c"</em>Code language: R (r)Solution:
This is of course possible without additional packages, but still difficult to read:
l$a[[1]]$b[3]
<em>## [1] "c"</em>Code language: R (r)pluck() from the purrr package, on the other hand, solves the task very smartly and easily understandable. The name or index of each level of the list is simply passed sequentially as an argument to the function:
l |> purrr::pluck("a", 1, "b", 3)
<em>## [1] "c"</em>Code language: R (r)4. rownames_to_column & rowid_to_column
Problem 1:
The row names of a dataset should be written to the first column. As an example we choose the well known mtcars dataset. In this record the row names describe the model of the car, which should be added to a new model column:
mtcars |> head()| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mazda RX4 | 21.0 | 6 | 160 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| Mazda RX4 Wag | 21.0 | 6 | 160 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| Datsun 710 | 22.8 | 4 | 108 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
| Hornet 4 Drive | 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| Hornet Sportabout | 18.7 | 8 | 360 | 175 | 3.15 | 3.440 | 17.02 | 0 | 0 | 3 | 2 |
| Valiant | 18.1 | 6 | 225 | 105 | 2.76 | 3.460 | 20.22 | 1 | 0 | 3 | 1 |
Solution:
The tibble package provides the function rownames_to_column(). The parameter var can be passed a string with the desired new column name. The new column is automatically placed at the first position of the record.
mtcars_model <- mtcars |> tibble::rownames_to_column(var = "model")
mtcars_model |> head()Code language: R (r)| model | mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mazda RX4 | 21.0 | 6 | 160 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| Mazda RX4 Wag | 21.0 | 6 | 160 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| Datsun 710 | 22.8 | 4 | 108 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
| Hornet 4 Drive | 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| Hornet Sportabout | 18.7 | 8 | 360 | 175 | 3.15 | 3.440 | 17.02 | 0 | 0 | 3 | 2 |
| Valiant | 18.1 | 6 | 225 | 105 | 2.76 | 3.460 | 20.22 | 1 | 0 | 3 | 1 |
Problem 2:
The second step is to add an index column that uniquely identifies each observation by an ID. To do this, we simply number the rows and write the row numbers in the new column.
Lösung:
An obvious solution creates a new column using mutate() in combination with nrow() or dplyr::row_number() and sets it to the first position using relocate():
mtcars_model |>
<em># alternativ: mutate(index = row_number()) |></em>
mutate(index = 1:nrow(mtcars)) |>
relocate(index) |>
head()Code language: R (r)| index | model | mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mazda RX4 | 21.0 | 6 | 160 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| 2 | Mazda RX4 Wag | 21.0 | 6 | 160 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| 3 | Datsun 710 | 22.8 | 4 | 108 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
| 4 | Hornet 4 Drive | 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| 5 | Hornet Sportabout | 18.7 | 8 | 360 | 175 | 3.15 | 3.440 | 17.02 | 0 | 0 | 3 | 2 |
| 6 | Valiant | 18.1 | 6 | 225 | 105 | 2.76 | 3.460 | 20.22 | 1 | 0 | 3 | 1 |
Again, the tibble package provides a more condensed solution. rowid_to_column() completes our task in one step. As before, the var argument can be used to specify the name of the new column:
mtcars_model |>
tibble::rowid_to_column(var = "index") |>
head()Code language: R (r)| index | model | mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mazda RX4 | 21.0 | 6 | 160 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| 2 | Mazda RX4 Wag | 21.0 | 6 | 160 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| 3 | Datsun 710 | 22.8 | 4 | 108 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
| 4 | Hornet 4 Drive | 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| 5 | Hornet Sportabout | 18.7 | 8 | 360 | 175 | 3.15 | 3.440 | 17.02 | 0 | 0 | 3 | 2 |
| 6 | Valiant | 18.1 | 6 | 225 | 105 | 2.76 | 3.460 | 20.22 | 1 | 0 | 3 | 1 |
5. parse_number
Solution:
In our daily work with data, we often encounter data sets that need to be cleaned up before they can be reused.
The following data set contains a column with products and another column with associated prices. However, the prices are included in a string without a fixed structure:
data_prices <- tibble(
product = 1:3,
costs = c("$10 -> expensive", "cheap: $2.50", "free, $0 !!")
)
data_pricesCode language: R (r)| product | costs |
|---|---|
| 1 | $10 -> expensive |
| 2 | cheap: $2.50 |
| 3 | free, $0 !! |
The task now is to separate from the strings the numerical prices for each product.
Solution:
A working, but often inconvenient, solution is to use regular expressions. In this example, we look for the first match of at least one digit followed by optional period and decimal places. A disadvantage of this approach is that the result column is still of type character:
one_or_more_digits <- "\d+"
optional_dot <- "\.?"
optional_digits <- "\d*"
data_prices |> mutate(price = stringr::str_extract(
string = costs,
pattern = paste0(one_or_more_digits, optional_dot, optional_digits)
))
<em>## # A tibble: 3 × 3</em>
<em>## product costs price</em>
<em>## <int> <chr> <chr></em>
<em>## 1 1 $10 -> expensive 10 </em>
<em>## 2 2 cheap: $2.50 2.50 </em>
<em>## 3 3 free, $0 !! 0</em>Code language: R (r)However, there is a more comfortable way: The readr package, which is usually used for data import, provides the helper function parse_number(). This scans a vector of strings for the first number and extracts it from its context. Possible decimal places are automatically taken into account.
The new price column in this case belongs directly to the double data type:
data_prices |> mutate(price = parse_number(costs))Code language: R (r)| product | costs | price |
|---|---|---|
| 1 | $10 -> expensive | 10.0 |
| 2 | cheap: $2.50 | 2.5 |
| 3 | free, $0 !! | 0.0 |
6. fct_lump_*
Problem:
In this example, we work with the babynames dataset from the R package of the same name, which lists the most popular baby names in the US over several decades. The column n indicates the absolute frequency of the name within a year:
babynames::babynames |> head()Code language: R (r)| year | sex | name | n | prop |
|---|---|---|---|---|
| 1880 | F | Mary | 7065 | 0.0723836 |
| 1880 | F | Anna | 2604 | 0.0266790 |
| 1880 | F | Emma | 2003 | 0.0205215 |
| 1880 | F | Elizabeth | 1939 | 0.0198658 |
| 1880 | F | Minnie | 1746 | 0.0178884 |
| 1880 | F | Margaret | 1578 | 0.0161672 |
We are interested in what the most frequent letters are for girls’ names to end in the year 2000:
names_2000 <- babynames::babynames |> filter(year == 2000)
last_letters_females <- names_2000 |>
mutate(last_letter = stringr::str_sub(name, start = -1, end = -1)) |>
filter(sex == "F") |>
count(last_letter, wt = n, name = "num_babies", sort = TRUE)
last_letters_females |> head(10)Code language: R (r)| last_letter | num_babies |
|---|---|
| a | 675963 |
| e | 318399 |
| n | 248450 |
| y | 246324 |
| h | 117324 |
| l | 56623 |
| r | 50769 |
| i | 42591 |
| s | 32603 |
| t | 9796 |
As expected, some letters are in last position much more often than others. For overview purposes, all letters with low frequency should be grouped into a common Other category.
Solution:
The forcats package helps us with this. The fct_lump_*() family aggregates rarer values of a factor (or here character) variable according to various criteria:
fct_lump_n()keeps the n most frequent values and merges all other values into a new category.fct_lump_min()summarizes all values which occur less often than a given absolute frequency.fct_lump_prop()summarizes all values which occur less often than a given relative frequency (proportion between 0 and 1).fct_lump_lowfreq()automatically summarizes the rarest values so that the aggregated Other category still has the lowest frequency among the new categories
In our example, we use fct_lump_n() and keep the most common last five letters:
last_letters_females_lumped <- last_letters_females |>
mutate(last_letter = factor(last_letter) |> fct_lump_n(
n = 5, w = num_babies, other_level = "Other"
)) |>
count(
last_letter,
wt = num_babies, name = "num_babies", sort = TRUE
)
last_letters_females_lumpedCode language: R (r)| last_letter | num_babies |
|---|---|
| a | 675963 |
| e | 318399 |
| n | 248450 |
| y | 246324 |
| Other | 208650 |
| h | 117324 |
The parameter w (for weight) can optionally specify a column whose values are summed up to determine the frequency. This is useful if, as in the example above, each letter occurs in only one line and the corresponding frequencies have already been calculated. The parameter is not needed if the frequencies have not yet been calculated and each letter would be duplicated n times in the last_letter column.
7. fct_reorder + geom_col
Problem:
Wir bleiben auch für dieses Beispiel bei dem babynames Datensatz und visualisieren die Anzahl der sechs häufigsten Mädchennamen in einem Balkendiagramm mit geom_col():
We also stick with the babynames dataset for this example and visualize the number of the six most common girls’ names in a bar chart using geom_col():
plot_color <- "#8bac37"
top_names_females <- names_2000 |>
filter(sex == "F") |>
slice_max(n, n = 6)
top_names_females |>
ggplot(aes(n, name)) +
geom_col(fill = plot_color) +
labs(
title = "Die 6 häufigsten Babynamen für Mädchen im Jahr 2000",
x = "Häufigkeit", y = NULL,
) +
theme_light() +
theme(plot.title = element_text(hjust = 0.5))Code language: R (r)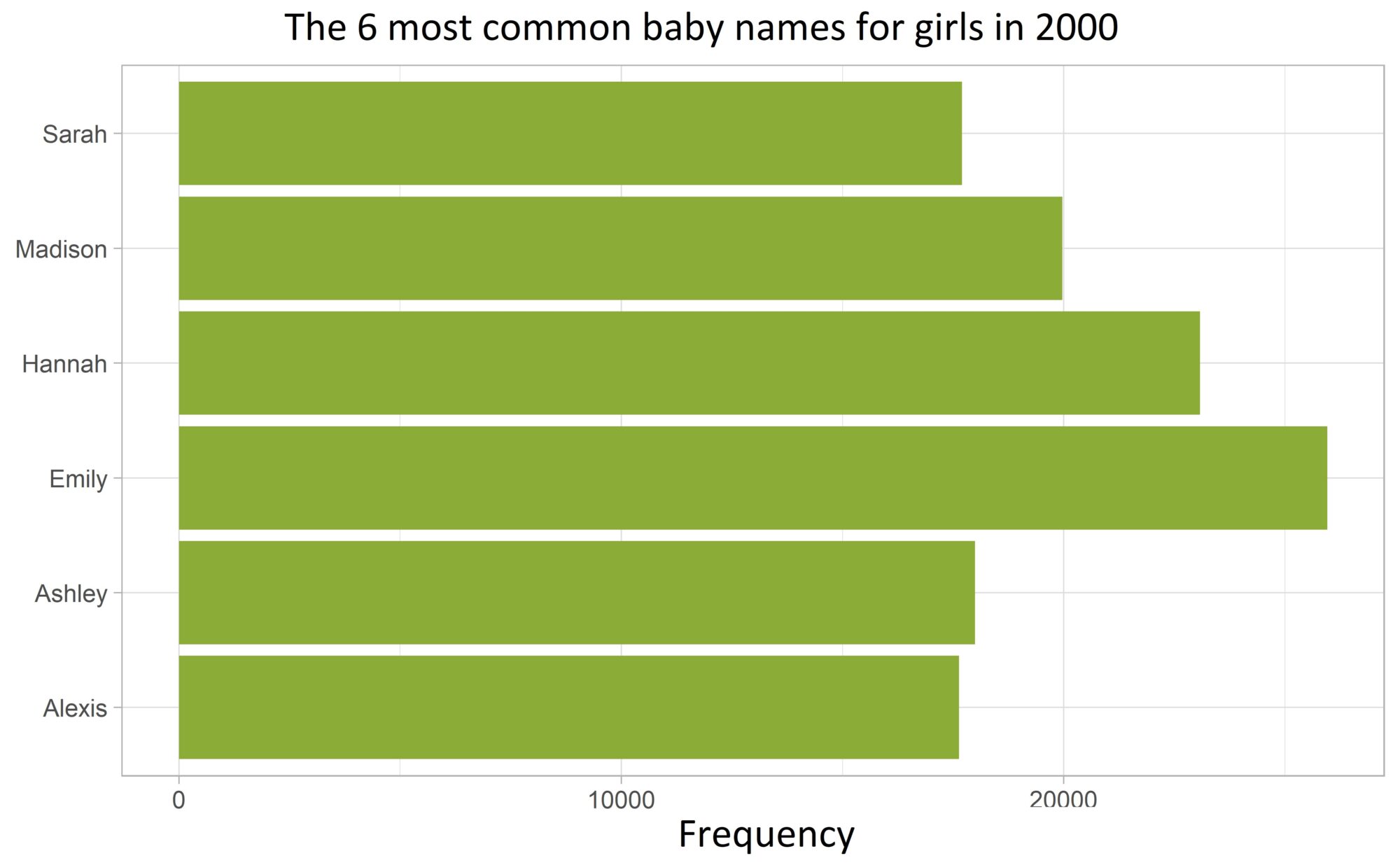
The names are not ordered along the y-axis according to their frequency!!
Solution:
To achieve this, we reorder the name column according to its frequency (of column n).
This case occurs so often in practice that I use geom_col() almost entirely in combination with fct_reorder() from the forcats package:
top_names_females |>
mutate(name = fct_reorder(name, n)) |>
ggplot(aes(n, name)) +
geom_col(fill = plot_color) +
labs(
title = "Die 6 häufigsten Babynamen für Mädchen im Jahr 2000",
x = "Häufigkeit", y = NULL,
) +
theme_light() +
theme(plot.title = element_text(hjust = 0.5))Code language: R (r)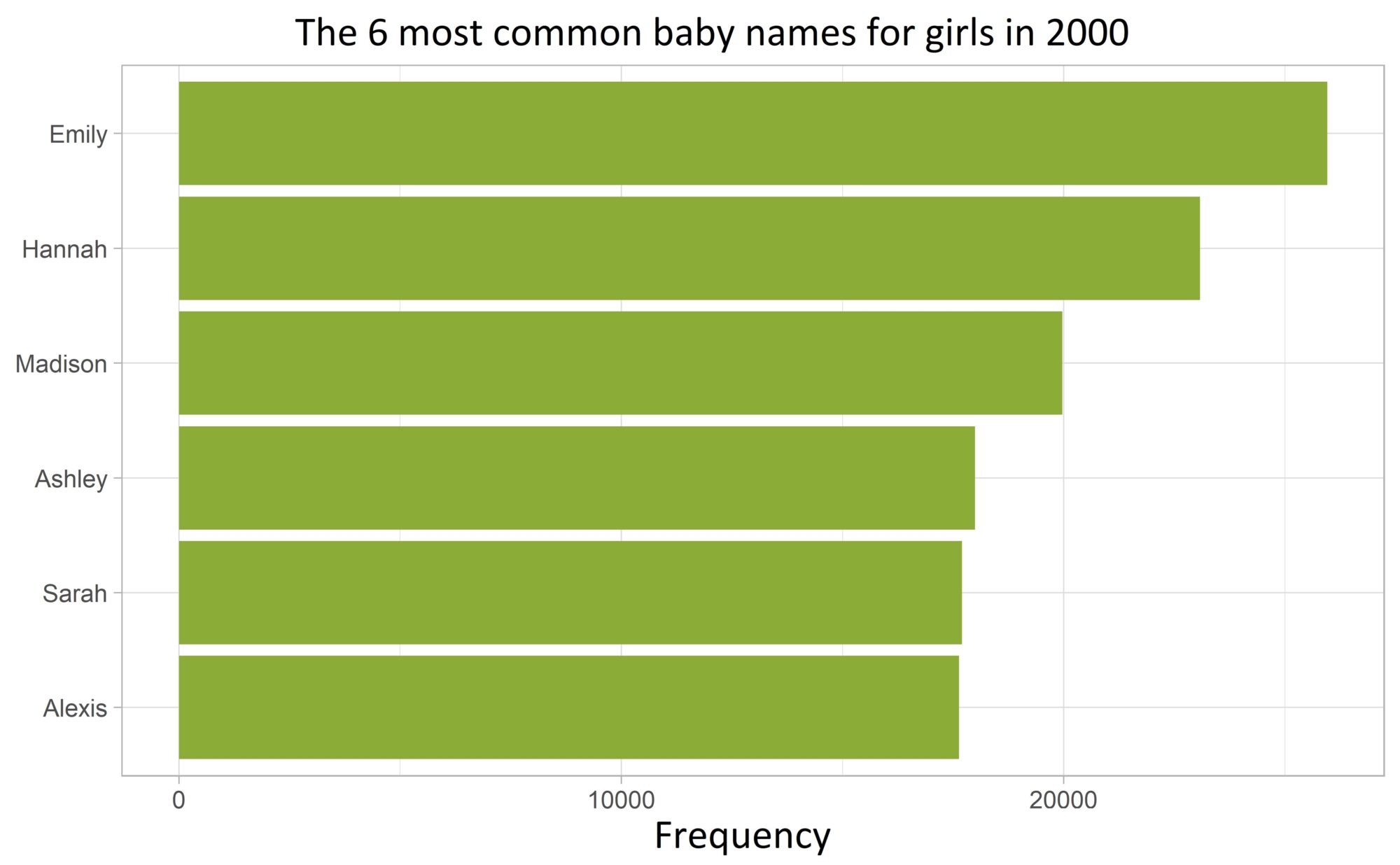
Bonus:
The above procedure no longer works as easily if a separate bar chart is to be plotted in descending frequency for each value of an additional factor variable. As an example, we now additionally consider the most frequent boy names:
top_names <- names_2000 |>
group_by(sex) |>
slice_max(n, n = 6)Code language: R (r)With fct_reorder(), the bars in each subplot are always ordered according to their frequency in the entire data set (and not just within each value of the sex variable).
The tidytext package, which is primarily used to analyze text data, saves us at this point.
The auxiliary functions reorder_within() and scale_y_reordered() do exactly the job and sort the values of the factor variables within each subplot:
top_names |>
mutate(name = tidytext::reorder_within(name, by = n, within = sex)) |>
ggplot(aes(n, name)) +
geom_col(fill = plot_color) +
labs(
title = "Die 6 häufigsten Babynamen für Mädchen und Jungs im Jahr 2000",
x = "Häufigkeit", y = NULL,
) +
facet_wrap(facets = vars(sex), scales = "free_y") +
tidytext::scale_y_reordered() +
theme_light() +
theme(plot.title = element_text(hjust = 0.5))Code language: R (r)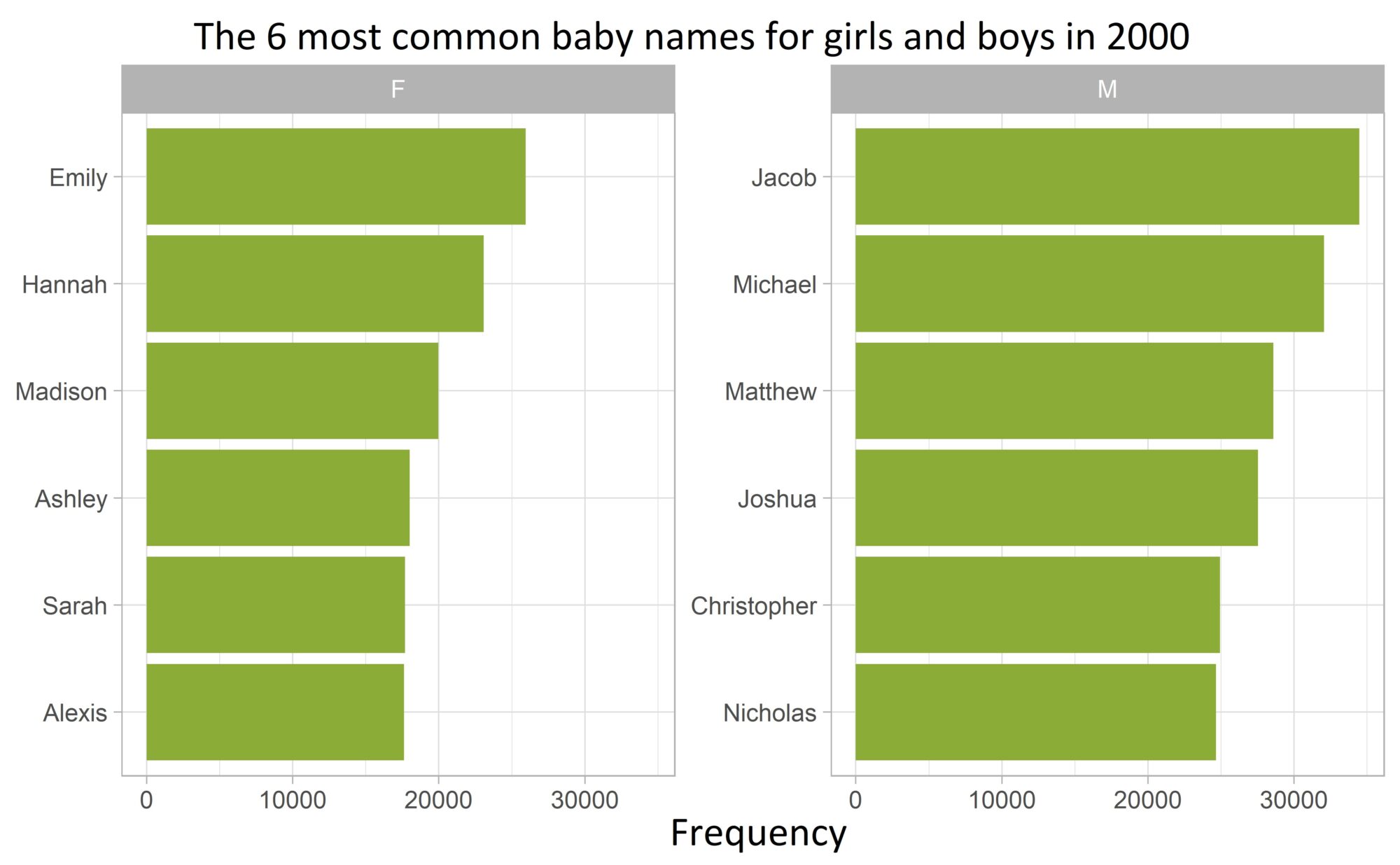
8. separate & separate_rows
Problem 1:
The following dataset should represent the results of different international soccer matches:
data_games <- tibble(
country = c("Germany", "France", "Spain"),
game = c("England - win", "Brazil - loss", "Portugal - tie")
)
data_gamesCode language: R (r)| country | game |
|---|---|
| Germany | England – win |
| France | Brazil – loss |
| Spain | Portugal – tie |
However, the game column includes two different types of information: the opponent as well as the result.
Solution:
To make the data frame tidy, we split the game column into two columns using the separate() function from the tidyr package:
data_games |> separate(col = game, into = c("opponent", "result"))Code language: R (r)| country | opponent | result |
|---|---|---|
| Germany | England | win |
| France | Brazil | loss |
| Spain | Portugal | tie |
Problem 2:
A similar problem occurs when a column contains two pieces of information of the same type in each row. The opponent column now includes only opposing teams, but several per row:
data_opponents <- tibble(
country = c("Germany", "France", "Spain"),
opponent = c("England, Switzerland", "Brazil, Denmark", "Portugal, Argentina")
)
data_opponentsCode language: R (r)| country | opponent |
|---|---|
| Germany | England, Switzerland |
| France | Brazil, Denmark |
| Spain | Portugal, Argentina |
In this case, the desired output does not contain more columns, but more rows, one for each opponent.
Solution:
separate_rows() splits each row of the opponent column into multiple rows, the corresponding country values are duplicated instead:
data_opponents |> separate_rows(opponent)Code language: R (r)| country | opponent |
|---|---|
| Germany | England |
| Germany | Switzerland |
| France | Brazil |
| France | Denmark |
| Spain | Portugal |
| Spain | Argentina |
9. str_flatten_comma
Problem:
A vector of strings is to be combined into a single string. All entries are separated by a comma, only the last two are to be connected by the connecting word “and”.
animals <- c("cat", "dog", "mouse", "elephant")
animals
<em>## [1] "cat" "dog" "mouse" "elephant"</em>Code language: R (r)Solution:
Without the stringr package, two calls to paste() are required:
- First, all entries except the last one are joined by a comma to form a single string.
- Then the result from step 1 is concatenated with the last vector entry.
paste(animals[-1], collapse = ", ") |> paste(animals[length(animals)], sep = " and ")
<em>## [1] "dog, mouse, elephant and elephant"</em>Code language: R (r)The stringr package provides its own function str_flatten_comma() with the very useful last parameter:
str_flatten_comma(animals, last = " and ")
<em>## [1] "cat, dog, mouse and elephant"</em>Code language: R (r)10. arrange + distinct
Problem:
The final example is inspired by work on a recent project of eoda. We have a dataset with two columns, the first column (group) contains an indicator for the group membership of each observation. Within each group, only a single row should be kept: The one with the highest numerical value in the second (value) column:
set.seed(123)
data_group_value <- tibble(
group = c(1, 3, 2, 1, 1, 2, 3, 1),
value = sample(1:100, size = 8, replace = FALSE)
)
data_group_valueCode language: R (r)| group | value |
|---|---|
| 1 | 31 |
| 3 | 79 |
| 2 | 51 |
| 1 | 14 |
| 1 | 67 |
| 2 | 42 |
| 3 | 50 |
| 1 | 43 |
Solution:
One possible approach is to use group_by() and slice_max() together:
data_group_value |>
group_by(group) |>
slice_max(value, n = 1)Code language: R (r)| group | value |
|---|---|
| 1 | 67 |
| 2 | 51 |
| 3 | 79 |
The disadvantage here is that for large datasets, a large number of groups may be formed, which reduces the efficiency of the calculation. In addition, this approach does not lead to the desired result for duplicates, since slice_max() selects all observations with the maximum value:
data_group_value_duplicates <- data_group_value |>
mutate(
value = case_when(
group == 1 ~ 20L,
TRUE ~ value
)
)
data_group_value_duplicates |>
group_by(group) |>
slice_max(value, n = 1)Code language: R (r)| group | value |
|---|---|
| 1 | 20 |
| 1 | 20 |
| 1 | 20 |
| 1 | 20 |
| 2 | 51 |
| 3 | 79 |
So in this case an additional call to slice(1) would be required to really keep only a single row per group.
A more efficient solution resorts to the dplyr combination of arrange() and distinct(). First, all rows within each group are sorted in descending order by their value values. The maximum value to be selected is therefore always at the first position within each group.
In the second step a call to distinct() is sufficient, because this function always keeps the first occurring value in case of duplicates and removes all others from the column
data_group_value_duplicates |>
arrange(group, desc(value)) |>
distinct(group, .keep_all = TRUE)Code language: R (r)| group | value |
|---|---|
| 1 | 20 |
| 2 | 51 |
| 3 | 79 |
Conclusion
In this article we have illustrated the usefulness of selected Tidyverse functions by means of various examples.
Some problems could be solved by other means as well
– but only with greater effort
Python, R & Shiny
Our trainings pave the way for your next steps. Machine Learning, Data Visualization, Time Series Analytics or Shiny:
Find the right course for your specific needs with us.